Blast
Description
The Basic Local Alignment Search Tool (BLAST) finds regions of local similarity between sequences. The program compares nucleotide or protein sequences to sequence databases and calculates the statistical significance of matches. BLAST can be used to infer functional and evolutionary relationships between sequences as well as help identify members of gene families. See Blast Program Selection Guide and the main blast website for more details.
Environment Modules
Run module spider blast to find out what environment modules are available for this application.
System Variables
- HPC_BLAST_DIR - installation directory
- BLASTDB - location of the blast databases.
Additional Information
We provide both NCBI BLAST+ and legacy blast binaries through the same modules.
BLAST databases
Follow the link to see the Information on the UFRC provided BLAST databases.
Job Script Examples
See the Blast_Job_Scripts page for blast Job script examples.
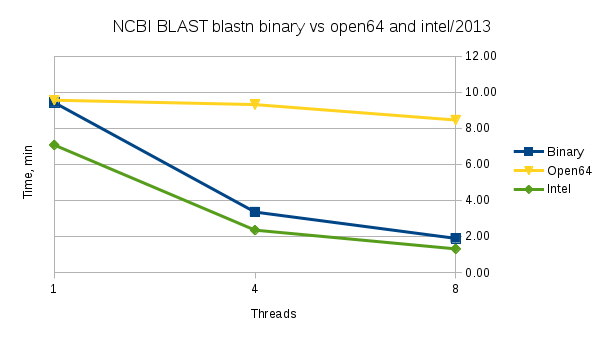
Performance
Performance comparison for the NCBI distributed binary 'blastn' vs binaries built by us from source using Intel's 2013 compiler or Open64 compiler. The open64 build behaves as a single-threaded build. The test data is a set of 2011229 nucleotide sequences used for the database and its subset of 1000 sequences is used as a query file.