Managing Python environments and Jupyter kernels: Difference between revisions
| Line 152: | Line 152: | ||
==== Start with <code>cudatoolkit</code> and </code>pytorch</code>/<code>tensorflow</code> if using GPU! ==== | ==== Start with <code>cudatoolkit</code> and </code>pytorch</code>/<code>tensorflow</code> if using GPU! ==== | ||
{{Note|'''If you plan on using a GPU''': it is important to both make the environment on a node with a GPU (within a Jupyter job for example) and to start by installing the <code>cudatoolkit</code> and <code>pytorch</code>, <code>tensorflow</code> or other frameworks. | {{Note|'''If you plan on using a GPU''': it is important to both make the environment on a node with a GPU (within a Jupyter job for example) and to start by installing the <code>cudatoolkit</code> and <code>pytorch</code>, <code>tensorflow</code> or other frameworks. <br> | ||
If you just <code>mamba install tensorflow</code>, you will get a version compiled with an older CUDA, which will be '''extremely''' slow or not recognize the GPU at all...ask me how I know 🤦. Same for <code>pytorch</code>.|warn}} | |||
From the [https://pytorch.org/get-started/locally/ PyTorch Installation page], we should use: | From the [https://pytorch.org/get-started/locally/ PyTorch Installation page], we should use: | ||
Revision as of 15:14, 29 November 2022
Background
Many projects that use Python code require careful management of the respective Python environments. Rapid changes in package dependencies, package version conflicts, deprecation of APIs (function calls) by individual projects, and obsolescence of system drivers and libraries make it virtually impossible to use an arbitrary set of packages or create one all-encompassing environment that will serve everyone's needs over long periods of time. The high velocity of changes in the popular ML/DL frameworks and packages and GPU computing exacerbates the problem.
The problem with pip install
Most guides and project documentation for installing python packages recommend using pip install for package installation. While pip is easy to use and works for many use cases, there are some major drawbacks. If you have spent any time working in Python, you will likely have seen (and may have run) suggestions to pip install ____, or within Jupyter !pip install ____, to install one or more packages. There are a few issues with doing pip install on a supercomputer like HiPerGator, though:
- Pip by default installs binary packages (wheels), which are often built on systems incompatible with HiPerGator. If you pip install a package and attempt to import it you might see an error about missing symbols or GLIBC version.
- Pip install of a package with no binary distribution (wheel) will attempt to build a package from source, but that build will likely fail without additional configuration.
- If you pip install a package that is already installed or will be later installed in an environment provided by UFRC, your version will take precedence over the packages installed in an environment provided by an environment module (or Jupyter kernel).
- Eventually package dependencies will become incompatible and you will encounter installation errors, import errors, or missing or wrong function calls (API changes). An innocuous
pip installof a single package can result in a drastic change of the environment rendering it unusable.
- Eventually package dependencies will become incompatible and you will encounter installation errors, import errors, or missing or wrong function calls (API changes). An innocuous
- Different packages may require different versions of the same package as dependencies leading to impossible to reconcile installation scenarios. This becomes a challenge to manage with
pipas there isn't a method to swap active versions. - On its own, `pip` installs **everything** in one location:
~/.local/lib/python3.X/site-packages/. All packages installed are in the same location for any given version of Python.
Conda and Mamba to the rescue!

conda and the newer, faster, drop-in replacement mamba, were written to solve some of these issues. They represent a higher level of packaging abstraction that can combine compiled packages, applications, and libraries as well as pip-installed python packages. They also allow easier management of project-specific environments and switching between environments as needed. They make it much easier to report the exact configuration of packages in an environment, facilitating reproducibility (recreation of an environment on a different system). Moreover, conda environments don't even have to be activated to be used. In most cases adding the path to the conda environment's 'bin' directory to the $PATH in the shell environment is sufficient for using them.
Check out the UFRC Help page on conda for additional information.
A caveat
conda and mamba get packages from channels, or repositories of prebuilt packages packages. While there are several available channels, like the main conda-forge, not every Python package is available from a conda channel as they have to be packaged for conda first. You may still need to use pip to install some packages as noted later. However, conda still helps manage environment by installing packages into separate directory trees rather than trying to install all packages into a single folder that pip does.
Getting Started: Conda Configuration
The ~/.condarc configuration file
conda's behavior is controlled by a configuration file in your home directory called .condarc. The dot at the start of the name means that the file is hidden from 'ls' file listing command by default. If you have not run conda before, you won't have this file. Whether the file exists or not, the steps here will help you modify the file to work best on HiPerGator. First load of the conda environment module on HiPerGator will put the current best practice .condarc into your home directory.
conda package cache location
conda caches (keeps a copy) of all downloaded packages by default in the ~/.conda/pkgs directory tree. If you install a lot of packages you may end up filling up your home quota. You can change the default package cache path. To do so, add or change the pkgs_dirs setting in your ~/.condarc configuration file e.g.:
pkgs_dirs: - /blue/mygroup/share/conda/pkgs
or
- /blue/mygroup/$USER/conda/pkgs
Replace mygroup with your actual group name.
conda environment location
conda puts all packages installed in a particular environment into a single directory. By default named conda environments are created in the ~/.conda/envs directory tree. They can quickly grow in size and, especially if you have many environments, fill the 40GB home directory quota. For example, the environment we will create in this training is 5.3GB in size. As such, it is important to use path based (conda create -p PATH) conda environments, which allow you to use any path for a particular environment for example allowing you to keep a project-specific conda environment close to the project data in /blue/ where you group has terrabyte(s) of space.
You can also change the default path for the name environments (conda create -n NAME) if you prefer to keep all conda environments in the same directory tree. To do so, add or change the envs_dirs setting in the ~/.condarc configuration file e.g.:
envs_dirs: - /blue/mygroup/share/conda/envs
or
- /blue/mygroup/$USER/conda/envs
Replace mygroup with your actual group name.
Expand this section to view instructions for editing your ~/.condarc file.
One way to edit your ~/.condarc file is to type: nano ~/.condarc`
If the file is empty, paste in the text below, editing the env_dirs: and pkg_dirs as below. If the file has contents, update those lines.
~/.condarc should look something like this when you are done editing (again, replacing group and user in the paths with your group and username).channels: - conda-forge - bioconda - defaults envs_dirs: - /blue/group/user/conda/envs pkgs_dirs: - /blue/group/user/conda/pkgs auto_activate_base: false auto_update_conda: false always_yes: false show_channel_urls: false
Create your first environment
Load the conda module
Before we can run conda or mamba on HiPerGator, we need to load the conda module:
module load conda
Create your first environment
Create a name based environment
To create your first name based (see path based instructions below)conda environment, run the following command. In this example, I am creating an environment named hfrl:
mamba create -n hfrl
The screenshot to the right is the output from running that command. Yours should look similar.
~/.condarc file. mamba will take care of that for you.Create a path based environment
To create a path based conda environment use the '-p PATH' argument:
mamba create -p PATH
e.g.
mamba create -p /blue/mygroup/share/project42/conda/envs/hfrl/
Activate the new environment
To activate our environment (whether created with mamba or conda we use the conda activate env_name command. Let's activate our new environment:
conda activate hfrl
or
conda activate /blue/mygroup/share/project42/conda/envs/hfrl/
Notice that your command prompt changes when you activate an environment to indicate which environment is active, showing that in parentheses before the other information:
(hfrl) [magitz@c0907a-s23 magitz]$
bin directory to $PATH in your job script.Install packages into our environment with mamba install
Now we are ready to start adding things to our environment.
There are a few ways to do this. We can install things one-by-one with either mamba install ____ or pip install ____. We will look at using yaml files below.
pip install will install the package into that environment. So, even if you continue using pip, adding conda environments solves the problem of everything being installed in one location--each environment has its own site-packages folder and is isolated from other environments.mamba install packages
Now we are ready to install packages using mamba install ___.
Start with cudatoolkit and pytorch/tensorflow if using GPU!
cudatoolkit and pytorch, tensorflow or other frameworks. If you just
mamba install tensorflow, you will get a version compiled with an older CUDA, which will be extremely slow or not recognize the GPU at all...ask me how I know 🤦. Same for pytorch.From the PyTorch Installation page, we should use:
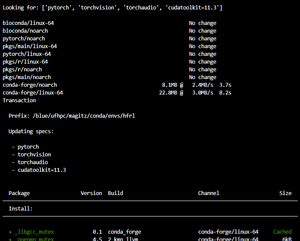
mamba install pytorch torchvision torchaudio cudatoolkit=11.3 -c pytorch
When you run that command, mamba will look in the repositories for the specified packages and their dependencies. Note we are specifying a particular version of cudatoolkit. As of May, 2022, that is the correct version on HiPerGator.
mamba will list the packages it will install and ask you to confirm the changes. Typing 'y' or hitting return will proceed; 'n' will cancel:
Finally, mamba will summarize the results:
Tensorflow installation alternative
While not needed for this tutorial, many users will want TensorFlow instead of PyTorch, so we will provide the command for that here. To install TensorFlow, use this command:
mamba install tensorflow cudatoolkit=11.2
This post at conda-forge has additional information and tips for installing particular versions or installing on a non-GPU node: GPU enabled TensorFlow builds on conda-forge.
Install additional packages
This tutorial creates an environment for the Hugging Face Deep Reinforcement Learning Course, you can either follow along with that or adapt to your needs.
You can list more than one package at a time in the mamba install command. We need a couple more, so run:
mamba install gym-box2d stable-baselines3
Add packages to our environment with pip install
As noted above, not everything is available in a conda channel. For example the next thing we want to install is huggingface_sb3.
If we type mamba install huggingface_sb3, we get a message saying nothing provides it as seen to the right:
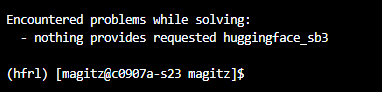
If we know of a conda source that has that package, we can add it to the channels: section of our ~/.condarc file. That will prompt mamba to include that location when searching.
But many things are only available via pip. So...
pip install huggingface_sb3
That will install huggingface_sb3. Again, because we are using environments and have the hfrl environment active, pip will not install huggingface_sb3 in our ~/.local/lib/python3.X/site-packages/ directory, but rather within in our hfrl directory, at /blue/group/user/conda/envs/hfrl/lib/python3.10/site-packages. This prevents the issues and headaches mentioned at the start.
Install additional packages
As with mamba, we could list multiple packages in the pip install command, but again, we only need one more:
pip install ale-py==0.7.4
Use your kernel from command line or scripts
Now that we have our environment ready, we can use it from the command line or a script using something like:
module load conda conda activate hfrl # Run my amazing python script python amazing_script.py
or with path based environments:
# Set path to environment we want and pre-pend to PATH variable env_path=/blue/mygroup/share/project42/conda/bin export PATH=$env_path:$PATH # Run my amazing python script python amazing_script.py
Setup a Jupyter Kernel for our environment
Often, we want to use the environment in a Jupyter notebook. To do that, we can create our own Jupyter Kernel.
Add the jupyterlab package
In order to use an environment in Jupyter, we need to make sure we install the jupyterlab package in the environment:
mamba install jupyterlab
Copy the template_kernel folder to your path
On HiPerGator, Jupyter looks in two places for kernels when you launch a notebook:
/apps/jupyterhub/kernels/for the globally available kernels that all users can use. (Also a good place to look for troubleshooting getting your own kernel going)~/.local/share/jupyter/kernelsfor each user. (Again, your home directory and the.localfolder is hidden since it starts with a dot)
Make the ~/.local/share/jupyter/kernels directory: mkdir -p ~/.local/share/jupyter/kernels
Copy the /apps/jupyterhub/template_kernel folder into your ~/.local/share/jupyter/kernels directory:
cp -r /apps/jupyterhub/template_kernel/ ~/.local/share/jupyter/kernels/hfrl
/apps/jupyterhub/kernels/ directory.Edit the template_kernel files
The template_kernel directory has four files: the run.sh and kernel.json files will need to be edited in a text editor. We will use nano in this tutorial. The logo-64X64.png and logo-32X32.png are icons for your kernel to help visually distinguish it from others. You can upload icons of those dimensions to replace the files, but they need to be named with those names.
Edit the kernel.json file
Let's start editing the kernel.json file. As an example, we can use:
nano ~/.local/share/jupyter/kernels/hfrl/kernel.json
The template has most of the information and notes on what needs to be updated. Edit the file to look like:
{
"language": "python",
"display_name": "HF_Deep_RL",
"argv": [
"~/.local/share/jupyter/kernels/hfrl/run.sh",
"-f",
"{connection_file}"
]
}
Edit the run.sh file
The run.sh file needs the path to the python application that is in our environment. The easiest way to get that is to make sure the environment is activated and run the command: which python
The path it outputs should look something like: /blue/group/user/conda/envs/hfrl/bin/python. Copy that path.
Edit the run.sh file with nano:
nano ~/.local/share/jupyter/kernels/hfrl/run.sh
The file should looks like this, but with your path:
#!/usr/bin/bash exec /blue/ufhpc/magitz/conda/envs/hfrl/bin/python -m ipykernel "$@"
If you are doing this in a Jupyter session, refresh your page. If not, launch Jupyter.
Your kernel should be there ready for you to use!
Working with yml files
Export your environment to an environment.yml file
Now that you have your environment working, you may want to document its contents and/or share it with others. The environment.yml file defines the environment and can be used to build a new environment with the same setup.
To export an environment file from an existing environment, run:
conda env export > hfrl.yml
You can inspect the contents of this file with cat hfrl.yml. This file defines the packages and versions that make up the environment as it is at this point in time. Note that it also includes packages that were installed via pip.
Create an environment from a yaml file
If you share the environment yaml file created above with another user, they can create a copy of your environment using the command:
conda env create --file hfrl.yml
They may need to edit the last line to change the location to match where they want their environment created.
Group environments
It is possible to create a shared environment accessed by a group on HiPerGator, storing the environment in, for example, /blue/group/share/conda. In general, this works best if only one user has write access to the environment. All installs should be made by that one user and should be communicated with the other users in the group.