Getting Started: Difference between revisions
Moskalenko (talk | contribs) |
Moskalenko (talk | contribs) No edit summary |
||
| Line 31: | Line 31: | ||
To use HiPerGator or HiPerGator-AI you need three basic parts | To use HiPerGator or HiPerGator-AI you need three basic parts | ||
* | * Interfaces | ||
You use Interfaces to interact with the system, manage data, initialize computation, and view the results. The main categories of interfaces 'Command-Line' also known as Terminal, Graphical User Interfaces, and Web Interfaces or applications for more specialized use. Some distinctions here are blurred because, for example, you can open a Terminal while using a Web Interface like [[JupyterHub]] or [[Open OnDemand]], but mostly you use a command-line Terminal interface through SSH connections (see below). | You use Interfaces to interact with the system, manage data, initialize computation, and view the results. The main categories of interfaces 'Command-Line' also known as Terminal, Graphical User Interfaces, and Web Interfaces or applications for more specialized use. Some distinctions here are blurred because, for example, you can open a Terminal while using a Web Interface like [[JupyterHub]] or [[Open OnDemand]], but mostly you use a command-line Terminal interface through SSH connections (see below). | ||
* | * Data Management | ||
To perform research analyses you need to [[Transfer_Data|upload]] and [[Storage|manage]] data. Note that misuse of the storage systems is the second main reason for account suspension after running analyses on login nodes. | To perform research analyses you need to [[Transfer_Data|upload]] and [[Storage|manage]] data. Note that misuse of the storage systems is the second main reason for account suspension after running analyses on login nodes. | ||
* | *Computation | ||
'''Warning:''' do not run full-scale (normal) analyses on login nodes. [[Development and Testing]] is required reading. The main approach to run computational analyses is through writing [[Sample SLURM Scripts|job scripts]] and sending them to the [[SLURM_Commands|scheduler]] to run. Some interfaces like [[Open OnDemand]], [[JupyterHub], and [[Galaxy]] can manage job scheduling behind the scenes and may be more convenient than job submission from the command-line when appropriate. | |||
==Interfaces== | ==Interfaces== | ||
| Line 81: | Line 82: | ||
</div> | </div> | ||
==Data Management== | ==Data Management== | ||
===Transferring Data=== | ===Transferring Data=== | ||
If you need to transfer datasets to or from HiPerGator and your local computer or another external location you have to pick the appropriate transfer mechanism. | If you need to transfer datasets to or from HiPerGator and your local computer or another external location you have to pick the appropriate transfer mechanism. | ||
| Line 100: | Line 102: | ||
===Globus=== | ===Globus=== | ||
Globus is a high-performance mechanism for file transfer. Globus works especially well for transferring large files or data sets | Globus is a high-performance mechanism for file transfer. Globus works especially well for transferring large files or data sets | ||
* [[Globus|See the Globus page]] for setup and configuration information. | * [[Globus|See the Globus page]] for setup and configuration information. | ||
| Line 122: | Line 123: | ||
You can also use your favorite file editor on your local machine, and then transfer the files to the cluster afterward. A caveat to this is that files created on Windows machines usually contain unprintable characters, which may be misinterpreted by Linux command interpreters (shells). If this happens, there is a utility called <code>dos2unix</code> that you can use to convert the text file from DOS/Windows formatting to Linux formatting. | You can also use your favorite file editor on your local machine, and then transfer the files to the cluster afterward. A caveat to this is that files created on Windows machines usually contain unprintable characters, which may be misinterpreted by Linux command interpreters (shells). If this happens, there is a utility called <code>dos2unix</code> that you can use to convert the text file from DOS/Windows formatting to Linux formatting. | ||
==Computation== | |||
==Using installed software== | ==Using installed software== | ||
The full list of software available for use can be viewed on the [[Installed_Software|Installed Software]] page. Access to installed software is provided through [[Modules|Environment Modules]]. | The full list of software available for use can be viewed on the [[Installed_Software|Installed Software]] page. Access to installed software is provided through [[Modules|Environment Modules]]. | ||
Revision as of 22:14, 14 January 2022
Welcome to UF Research Computing! This page is intended to help new clients get started on HiPerGator.
From Zero to HiPerGator
Initial Consult
If a face-to-face discussion about the group's needs is needed you can meet one of the UF Research Computing Facilitators face-to-face or virtually or submit a support request to start the conversation.
HiPerGator Accounts
Group's sponsor has to be the first person to request a HiPerGator account indicating that they are a new sponsor. In the process we will create their sponsored group.
Afterwards, group members will be able to submit HiPerGator account requests indicating their PI as the sponsor. Once approved, their linux accounts will be created.
Trial Allocation
We recommend that the group's sponsor requests a free trial allocation for storage and computational resources to get the group started on HiPerGator. Group members can then use HiPerGator for the 3 month duration of the trial allocation to figure out what resources and applications they really need.
Purchasing Resources
After or while the group uses a trial allocation to determine the computational and storage resources it needs the group's sponsor can submit a purchase request for hardware (5-years) or services (3-months to longer) to invest into the resources to cover the group's HiPerGator use.
Some groups may have access to shared departmental allocations. In this case, instead of purchasing resources, group members can request to be added to the departmental group to gain access to the shared resources.
Some examples of departments with shared allocations include the Genetics Institute, Emerging Pathogens Institute, Statistics Department, Biostatistics Department, Center for Compressible Multiphase Turbulence (CCMT), Cognitive Aging and Memory Clinical Translational Research Program (CAMCTRP), Center for Molecular Magnetic Quantum Materials, Physics Department, and Plant Pathology Department. In addition, several research groups working on collaborative projects have shared allocations accessible to members of those projects.
At this point a group is established on HiPerGator and can continue their computational work. See below for more details on the basic use.
Introduction to Using HiPerGator
- Note
- see a short Quick Start Guide for some hints on getting going and avoiding common pitfalls.
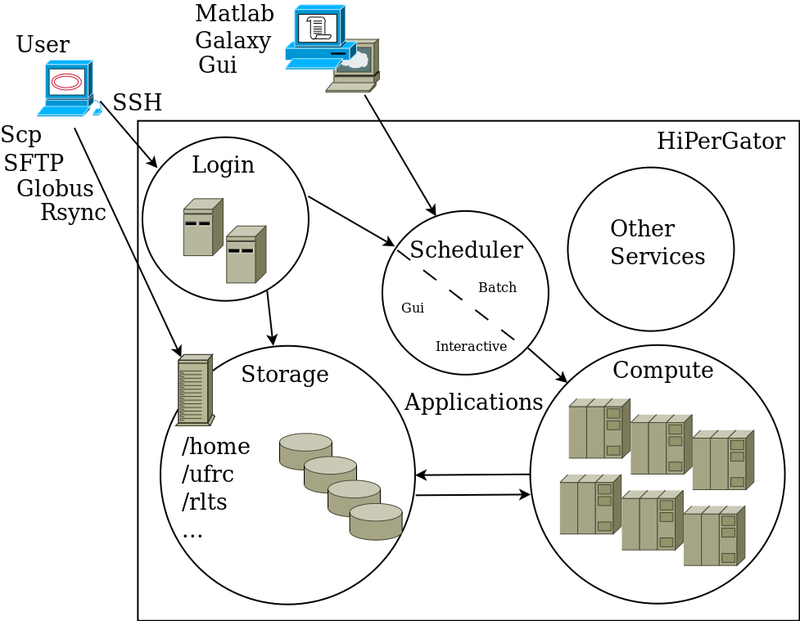
To use HiPerGator or HiPerGator-AI you need three basic parts
- Interfaces
You use Interfaces to interact with the system, manage data, initialize computation, and view the results. The main categories of interfaces 'Command-Line' also known as Terminal, Graphical User Interfaces, and Web Interfaces or applications for more specialized use. Some distinctions here are blurred because, for example, you can open a Terminal while using a Web Interface like JupyterHub or Open OnDemand, but mostly you use a command-line Terminal interface through SSH connections (see below).
- Data Management
To perform research analyses you need to upload and manage data. Note that misuse of the storage systems is the second main reason for account suspension after running analyses on login nodes.
- Computation
Warning: do not run full-scale (normal) analyses on login nodes. Development and Testing is required reading. The main approach to run computational analyses is through writing job scripts and sending them to the scheduler to run. Some interfaces like Open OnDemand, [[JupyterHub], and Galaxy can manage job scheduling behind the scenes and may be more convenient than job submission from the command-line when appropriate.
Interfaces
Connecting to a HiPerGator Terminal via SSH
To work on HiPerGator you will have to connect to it from your local computer either via SSH (terminal session) or via one of the web/application interfaces we provide such as Galaxy, Open OnDemand, or JupyterHub.
For any given command below, <username> should be replaced with the UFRC username (same as your GatorLink username).
Connecting from Windows
Expand this section to view instructions for logging in with Windows.
Connecting from Linux and MacOS
Expand to view instructions for connecting from Linux or MacOS.
Data Management
Transferring Data
If you need to transfer datasets to or from HiPerGator and your local computer or another external location you have to pick the appropriate transfer mechanism.
SFTP=
SFTP, or secure file transfer, works well for small to medium data transfers and is appropriate for both small and large data files.
If you would like to use a Graphical User Interface secure file transfer client we recommend:
FileZilla for Windows or MacOS X.
- The FileZilla installer contains adware/malware and probably should be avoided.
After you have chosen and downloaded a client, configure the client to connect to hpg.rc.ufl.edu, specifying port number 22. Use your username and password to log in.
Rsync
If you prefer to use the command-line or to get maximum efficiency from your data transfers Rsync, which is an incremental file transfer utility that minimizes network usage, is a good choice. It does so by transmitting only the differences between local and remote files rather than transmitting complete files every time a sync is run as SFTP does. Rsync is best used for tasks like synchronizing files stored across multiple subdirectories, or updating large data sets. It works well both for small and large files. See the Rsync page for instructions on using rsync.
Globus
Globus is a high-performance mechanism for file transfer. Globus works especially well for transferring large files or data sets
- See the Globus page for setup and configuration information.
Samba
Samba service, also known as a 'network share' or 'mapped drive' provides you with an ability to connect to some HiPerGator filesystems as locally mapped drives (or mount points on Linux or MacOS X).Once you connected to a share this mechanism provides you with a file transfer option that allows you to use your client computer's native file manager to access and manage your files. UFRC Samba setup does not provide high performance, so try to use it sparingly and for smaller files, like job scripts or analysis reports to be copied to your local system. You must be connected to the UF network (either on-campus or through the VPN) to connect to Samba shares.
- See the page on accessing Samba for setup information specific to your computer's operating system.
Automounted Paths
Note: NFS-based storage on our systems are typically automounted, which means they are dynamically mounted only when users are actually accessing them. For example if you have an invested folder as /orange/smith, to access it you will have to specifically type in the full path of "/orange/smith" to be able to see the contents and access them. Directly browsing /orange will not show the smith sub-folder unless someone else is using it coincidentally. Automounted folders are pretty common on the systems, they include /orange, /bio, /rlts and even /home etc.
Editing your files
Several methods exist for editing your files on the cluster.
Native Editors
- vi - The visual editor (vi) is the traditional Unix editor; however, it is not necessarily the most intuitive editor. View a tutorial for using vi
- emacs - Emacs is a much heavier duty editor, but again has the problem of having commands that are non-intuitive. View a tutorial for using emacs
- pico - While pico is not installed on the system, nano is installed, and is a pico work-a-like.
- nano - Nano has a good bit of on-screen help to make it easier to use.
External Editors
You can also use your favorite file editor on your local machine, and then transfer the files to the cluster afterward. A caveat to this is that files created on Windows machines usually contain unprintable characters, which may be misinterpreted by Linux command interpreters (shells). If this happens, there is a utility called dos2unix that you can use to convert the text file from DOS/Windows formatting to Linux formatting.
Computation
Using installed software
The full list of software available for use can be viewed on the Installed Software page. Access to installed software is provided through Environment Modules.
The following command can be used to browse the full list of available modules, along with short descriptions of the applications they make available:
module spider
To load a module, use the following command:
module load <module_name>
For more information on loading modules to access software, view the page on the basic usage of environment modules.
There are some useful commands and utilities in a 'ufrc' environment module in addition to installed applications.
Doing Interactive Testing or Development
You don't always have to use the SLURM scheduler. When all you need is a quick shell session to run a command or two, write and/or test a job script, or compile some code use SLURM Dev Sessions.
Running Graphical Programs
It is possible to run programs that use a graphical user interface (GUI) on the system. However, doing so requires an installation of and configuration of additional software on the client computer.
Please see the GUI Programs page for information on running graphical user interface applications at UFRC.
Scheduling computational jobs
UFRC uses the Simple Linux Utility for Resource Management, or SLURM, to allocate resources and schedule jobs. Users can create SLURM job scripts to submit jobs to the system. These scripts can, and should, be modified in order to control several aspects of your job, like resource allocation, email notifications, or an output destination.
- See the Annotated SLURM Script for a walk-through of the basic components of a SLURM job script
- See the Sample SLURM Scripts for several SLURM job script examples
To submit a job script from one of the login nodes accessed via hpg.rc.ufl.edu, use the following command:
$ sbatch <your_job_script>
To check the status of submitted jobs, use the following command:
$ squeue -u <username>
View SLURM_Commands for more useful SLURM commands.
Managing Cores and Memory
The amount of resources within an investment is calculated in NCU (Normalized Computing Units), which correspond to 1 CPU core and about 3.5GB of memory for each NCU purchased. CPUs (cores) and RAM are allocated to jobs independently as requested by your job script.
Your group's investment can run out of **cores** (SLURM may show QOSGrpCpuLimit in the reason a job is pending) OR **memory** (SLURM may show QOSGrpMemLimit in the reason a job is pending) depending on current use by running jobs.
The majority of HiPerGator nodes have the same ratio of about 4 GB of RAM per core, which, after accounting for the operating system and system services, leaves about 3.5 GB usable for jobs; hence the ratio of 1 core and 3.5GB RAM per NCU.
Most HiPerGator nodes have 32 cores and 128 GB RAM (~30,000 cores in the newer part of the cluster) or 64 cores and 256 GB RAM (~16,000 cores in the older part of the cluster). The bigmem nodes and the newer Skylake nodes have a higher ratio of 16 GB/core and 6 GB/core, respectively. See Available_Node_Features for the exact data on resources available on all types of nodes on HiPerGator.
You must specify both the number of cores and the amount of RAM needed in the job script for SLURM with the --mem (total job memory) or --mem-per-cpu (per-core memory) options. Otherwise, the job will be assigned the default 600mb of memory.
If you need more than 128 GB of RAM, you can only run on the older nodes, which have 256 GB of RAM, or on the bigmem nodes, which have up to 1.5 TB of RAM.
See Account and QOS limits under SLURM for an extensive explanation of QOS and SLURM account use.
Getting help
If you are having problems using the UFRC system, please let our staff know by submitting a support request.
Visual Overview
The diagram below shown a high-level overview of HiPerGator use. We will go over each part in sections below